Polymerase Chain Reaction (PCR), the technique to synthesize DNA, is the foundation of NGS
The key is during DNA synthesize, only add one nucleotide at a time, pause the reaction, record that base, then add & record another. The strength of NGS is massive parallelization - the ability to sequence billions of DNA at a time.
Sequencing by “eavesdropping” each step of DNA synthesis
- Imagine a DNA is a long Lego pillar like the picture below, where each small cubic building block is a nucleotide. 4 colors represent 4 types of bases AGCT.
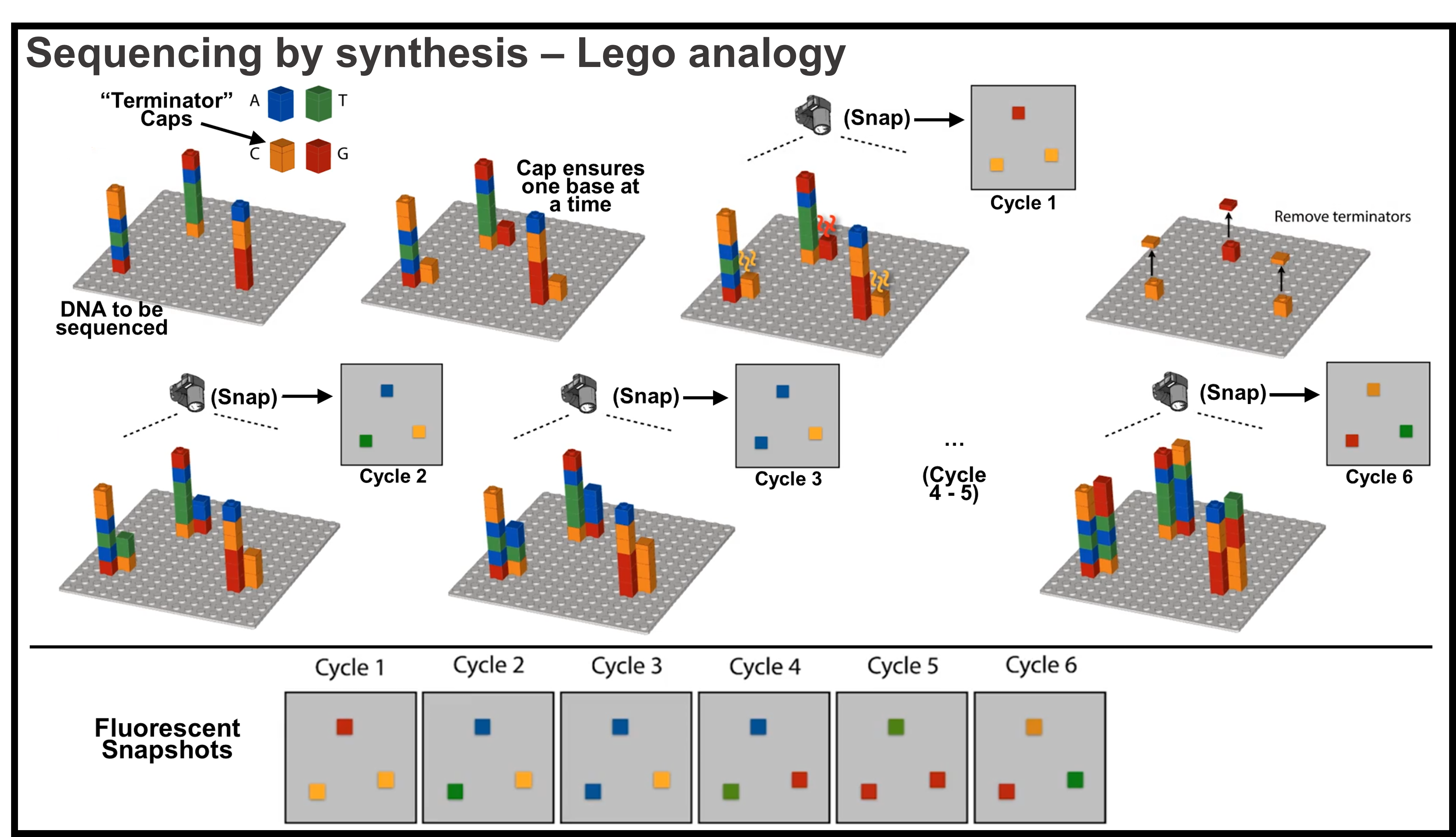
Image adapted from Coursera Course: Algorithms for DNA Sequencing
- Sequencing starts by attaching DNAs to be sequenced (called templates) to a surface. There are 3 DNA templates / Lego pillars in the toy system (shown at top left corner of the picture), while in reality billions of DNAs would be sequenced at a time.
- Then specially modified nucleotides AGCT are added as reaction materials to synthesize/duplicate DNAs. The nucleotides are special in two ways: they have a terminator “cap”; and they fluoresce.
- The cap prevents another nucleotides (AGCT Lego cubes) from being added on to the capped nucleotide. This allows the DNA molecule to grow one nucleotide/base at a time, to pause the reaction so the current base can be read. The cap can be removed after the signal is taken.
- For example, at cycle 1, the three DNAs pillars have a paired strand of new DNA being synthesized right next to them, because of the capped AGCT cubes, only one cube was added to the new strand (paired according to A-T, C-G rule).
- These newly added nucleotide cubes fluoresce - more importantly, different types of nucleotides fluoresce different colors, which allows us to read the base identity by their color.
- A camera at top takes the fluorescent snapshot, and we learned what base is added. For example, the snapshot of cycle 1 tells us a G (red) is added to the top DNA being synthesized. We can infer the DNA being sequenced (the paired strand) has a C at the same position.
- Once the snapshot is taken, the nucleotides are washed out, the caps are removed, opening up the Lego cube for new cubes to be added in the next cycle. (In the picture showing the caps being removed, the original DNA pillars are not shown, but they are still attached to the surface).
- The cycle repeats, one capped nucleotide is added in the next cycle and a snapshot taken, the identity of the second nucleotides are read.
Massive parallelization: billions of reads per run
- Billions of DNA can be sequenced at the same time in a parallel fashion, by attaching billions of the Lego pillars and recording their signals all at once.
- In reality, the size of one DNA molecule is too small (~nm size) for the fluorescent light to be detected. Each DNA is duplicated to a cluster of itself to generate enough signals for a sequencer/human to read. An example of sequencing snapshot and base calling in each cycle is shown below.
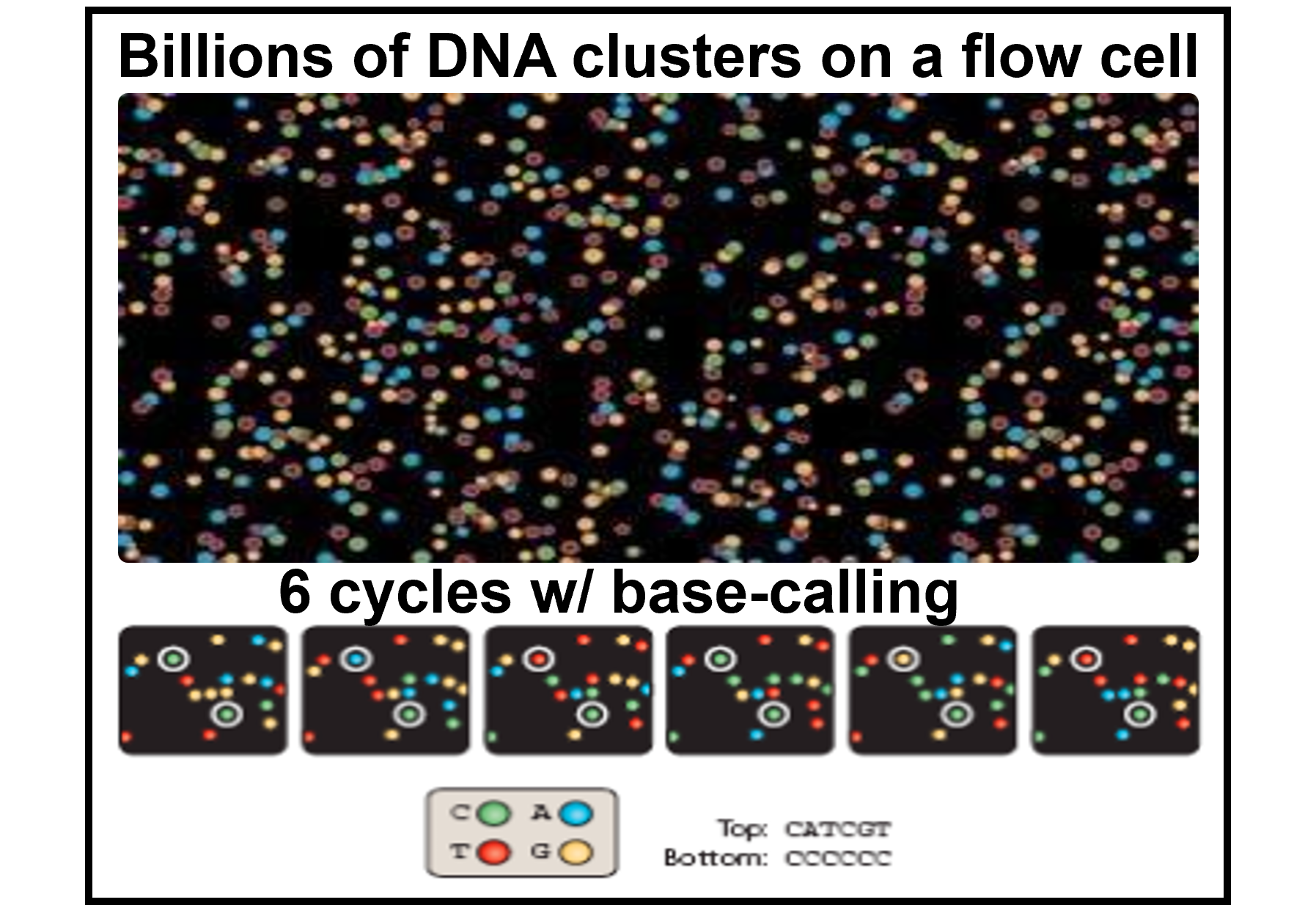 Image adapted from Applied Computational Genomics Course by Dr. Aaron Quinlan
Image adapted from Applied Computational Genomics Course by Dr. Aaron Quinlan
150bp read length and pair end sequencing
- NGS (Illumina) sequence DNA reads of ~150 base pair (bp) length (limited by errors such as fluorescence bleed through, more discussion later).
- 150bp is smaller than many repetitive segments of human genome, making them hard to place back (align) to the reference genome.
- One clever get around is prepare DNA samples of ~500 bp long, and sequence 75 bp on each end - pair end sequencing. An example is shown here, the first and last 75 bp of the sample are sequenced, with ~350 bp sequence in the middle missing.
5' - AACTTTAC(75bp)..../~350pb/....
..../~350pb/....TCCTTACA(75bp) - 3'
- The 75bp sequence from each end might align to thousands of loci (places) on the reference genome, but usually only one or two of them are within 500bp of each other.
Sequencing process
- All chromosomes from cell samples are shredded to ~500pb DNA strands (templates).
- Each DNA strand (template) is tethered to a slide, with billions of templates on the same slides.
- PCR duplicate each DNA template to a cluster of identical DNA strands, so that the fluorescence emitted by the cluster can be detected and read by sequencers.
- Add fluorescent labeled, terminator modified ATCG nucleotides. They will pair with the templates, and the pairing DNA strand extend only one base.
- Shine fluorescent light to read the current base.
- The terminator modification is reversed, and the pairing DNAs can be extended again, for the next nucleotide to be attached and read.
- Repeat until ideal length (150/75bp) of DNA is sequenced.
Sequencing error and base quality
- Out of sync bases
- Ideally the paired DNA adds one base at a time (on schedule), but some strands may miss a base (behind) or added more than one base (ahead). The out of sync bases can shine a different color and cause the signal to be less pure.
- The out of sync issue causes read quality to decrease in later cycles, and limiting read length to ~hundreds bp.
- Fluorescence bleed through between adjacent clusters
- Two clusters can get too close to each other, causing their fluorescence light/color to bleed over each other, making the color less pure and hard to identify.
- Quality score , where is the probability of the base identity is wrong.
- E.g. a base has 10% (P_err=0.1) chance of being a sequencing error, its quality score
- Rule of thumb is Q>30 (P_err=0.001) - error of 1 in a thousand, is a high quality nucleotide.
NGS Applications - Various Sequencing Types talks about what bio information we can extract by sequencing DNAs in specific settings.